Abstract
Cell function depends on tissue rigidity, which cells probe by applying and transmitting forces to their extracellular matrix, and then transducing them into biochemical signals. Here we show that in response to matrix rigidity and density, force transmission and transduction are explained by the mechanical properties of the actin–talin–integrin–fibronectin clutch. We demonstrate that force transmission is regulated by a dynamic clutch mechanism, which unveils its fundamental biphasic force/rigidity relationship on talin depletion. Force transduction is triggered by talin unfolding above a stiffness threshold. Below this threshold, integrins unbind and release force before talin can unfold. Above the threshold, talin unfolds and binds to vinculin, leading to adhesion growth and YAP nuclear translocation. Matrix density, myosin contractility, integrin ligation and talin mechanical stability differently and nonlinearly regulate both force transmission and the transduction threshold. In all cases, coupling of talin unfolding dynamics to a theoretical clutch model quantitatively predicts cell response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Moore, S. W., Roca-Cusachs, P. & Sheetz, M. P. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell 19, 194–206 (2010).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Chan, C. E. & Odde, D. J. Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008).
Elosegui-Artola, A. et al. Rigidity sensing and adaptation through regulation of integrin types. Nat. Mater. 13, 631–637 (2014).
Case, L. B. & Waterman, C. M. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Gupta, M. et al. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat. Commun. 6, 7525 (2015).
Oakes, P. W., Banerjee, S., Marchetti, M. C. & Gardel, M. L. Geometry regulates traction stresses in adherent cells. Biophys. J. 107, 825–833 (2014).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Ghibaudo, M. et al. Traction forces and rigidity sensing regulate cell functions. Soft Matter 4, 1836–1843 (2008).
Schiller, H. B. et al. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15, 625–636 (2013).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on sub-micrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012).
Califano, J. P. & Reinhart-King, C. A. Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact. Cell. Mol. Bioeng. 3, 68–75 (2010).
Etienne, J. et al. Cells as liquid motors: mechanosensitivity emerges from collective dynamics of actomyosin cortex. Proc. Natl Acad. Sci. USA 112, 2740–2745 (2015).
Roca-Cusachs, P., Iskratsch, T. & Sheetz, M. P. Finding the weakest link—exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125, 3025–3038 (2012).
Schoen, I., Pruitt, B. L. & Vogel, V. The Yin-Yang of rigidity sensing: how forces and mechanical properties regulate the cellular response to materials. Annu. Rev. Mater. Res. 43, 589–618 (2013).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Margadant, F. et al. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 9, e1001223 (2011).
Roca-Cusachs, P., Gauthier, N. C., del Rio, A. & Sheetz, M. P. Clustering of α5β1 integrins determines adhesion strength whereas αvβ3 and talin enable mechanotransduction. Proc. Natl Acad. Sci. USA 106, 16245–16250 (2009).
Zhang, X. et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 10, 1062–1068 (2008).
del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Chen, H., Choudhury, D. M. & Craig, S. W. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281, 40389–40398 (2006).
Wegener, K. L. et al. Structural basis of integrin activation by talin. Cell 128, 171–182 (2007).
Yao, M. et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 (2014).
Kong, F., Garcia, A. J., Mould, A. P., Humphries, M. J. & Zhu, C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009).
Roca-Cusachs, P. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl Acad. Sci. USA 110, E1361–E1370 (2013).
Tanentzapf, G. & Brown, N. H. An interaction between integrin and the talin FERM domain mediates integrin activation but not linkage to the cytoskeleton. Nat. Cell Biol. 8, 601–606 (2006).
Hirata, H., Tatsumi, H., Lim, C. T. & Sokabe, M. Force-dependent vinculin binding to talin in live cells: a crucial step in anchoring the actin cytoskeleton to focal adhesions. Am. J. Physiol. Cell Physiol. 306, C607–C620 (2014).
Cohen, D. M., Kutscher, B., Chen, H., Murphy, D. B. & Craig, S. W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 281, 16006–16015 (2006).
Humphries, J. D. et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 (2007).
Bangasser, B. L., Rosenfeld, S. S. & Odde, D. J. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 105, 581–592 (2013).
Bangasser, B. L. & Odde, D. J. Master equation-based analysis of a motor-clutch model for cell traction force. Cell. Mol. Bioeng. 6, 449–459 (2013).
Saltel, F. et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 187, 715–731 (2009).
Klapholz, B. et al. Alternative mechanisms for talin to mediate integrin function. Curr. Biol. 25, 847–857 (2015).
Iwamoto, D. V. & Calderwood, D. A. Regulation of integrin-mediated adhesions. Curr. Opin. Cell Biol. 36, 41–47 (2015).
Case, L. B. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17, 880–892 (2015).
Thievessen, I. et al. Vinculin–actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 (2013).
Roca-Cusachs, P. et al. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys. J. 94, 4984–4995 (2008).
Swift, J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013).
DuFort, C. C., Paszek, M. J. & Weaver, V. M. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319 (2011).
Plouffe, S. W., Hong, A. W. & Guan, K. L. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 21, 212–222 (2015).
Bate, N. et al. Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics. PLoS ONE 7, e34461 (2012).
Simonson, W. T., Franco, S. J. & Huttenlocher, A. Talin1 regulates TCR-mediated LFA-1 function. J. Immunol. 177, 7707–7714 (2006).
Pierschbacher, M. D. & Ruoslahti, E. Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding-specificity in cell-adhesion. J. Biol. Chem. 262, 17294–17298 (1987).
Frelinger, A. L. 3rd, Du, X. P., Plow, E. F. & Ginsberg, M. H. Monoclonal antibodies to ligand-occupied conformers of integrin α IIb β 3 (glycoprotein IIb-IIIa) alter receptor affinity, specificity, and function. J. Biol. Chem. 266, 17106–17111 (1991).
Alcaraz, J. et al. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys. J. 84, 2071–2079 (2003).
Hutter, J. L. & Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64, 1868–1873 (1993).
Serra-Picamal, X. et al. Mechanical waves during tissue expansion. Nat. Phys. 8, U628–U666 (2012).
Butler, J. P., Tolic-Norrelykke, I. M., Fabry, B. & Fredberg, J. J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282, C595–C605 (2002).
Elosegui-Artola, A. et al. Image analysis for the quantitative comparison of stress fibers and focal adhesions. PLoS ONE 9, e107393 (2014).
Chen, Y. et al. Fluorescence biomembrane force probe: concurrent quantitation of receptor-ligand kinetics and binding-induced intracellular signaling on a single cell. J. Vis. Exp. 102, e52975 (2015).
Takagi, J., Petre, B. M., Walz, T. & Springer, T. A. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–611 (2002).
Chesla, S. E., Selvaraj, P. & Zhu, C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 75, 1553–1572 (1998).
Chen, W., Zarnitsyna, V. I., Sarangapani, K. K., Huang, J. & Zhu, C. Measuring receptor-ligand binding kinetics on cell surfaces: from adhesion frequency to thermal fluctuation methods. Cell. Mol. Bioeng. 1, 276–288 (2008).
Chen, W., Lou, J. & Zhu, C. Forcing switch from short- to intermediate- and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285, 35967–35978 (2010).
Acknowledgements
We acknowledge support from the Spanish Ministry for Economy and Competitiveness (BFU2011-23111, BFU2012-38146 and BFU2014-52586-REDT), a Career Integration Grant within the seventh European Community Framework Programme (PCIG10-GA-2011-303848), the European Research Council (Grant Agreements 242993 and 240487), the Generalitat de Catalunya, Fundació La Caixa, Fundació la Marató de TV3 (project 20133330), and the National Institutes of Health (US NIH R01AI044902). A.E.-A., R.O. and C.P.-G. were supported respectively by a Juan de la Cierva Fellowship (Spanish Ministry of Economy and Competitiveness), a FI fellowship (Generalitat de Catalunya), and the fundació ‘La Caixa’. We thank R. Sunyer, J. Alcaraz, E. Bazellières, F. Rico, S. Garcia-Manyes, N. Bate, N. Berrow and the members of the P.R.-C. and X.T. laboratories for technical assistance and discussions.
Author information
Authors and Affiliations
Contributions
A.E.-A. and P.R.-C. conceived the study, A.E.-A., C.Z., X.T. and P.R.-C. designed the experiments, A.E.-A., R.O., Y.C., A.K., C.P.-G. and N.C. performed the experiments, P.R.-C. carried out the theoretical modelling, and A.E.-A. and P.R.-C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The results have been protected under a patent application.
Integrated supplementary information
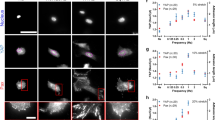
Supplementary Figure 4 Cell adhesion to fibronectin-coated gels is mediated by αvβ3 and α5β1 integrins.
(a) Images showing control and Talin 2 shRNA cells on 29 kPa fibronectin-coated polyacrylamide gels with or without blocking integrin α5β1 (using 10 μg ml−1 of BMB5 antibody), αvβ3 (using 0.5 mM of the specific Gpen peptide) or both. Scale bar is 50 μm. (b) Corresponding quantification of the percentage of spread cells (from left to right, n = 12, 12, 11, 11, 20, 11, 10, 12 fields of view)(∗∗∗, p ≤ 0.001, two-way Anova). Data show 1 out of 2 independent experiments. Blocking both integrins abolished cell adhesion almost completely.
Supplementary Figure 5 Talin1 Head L325R expression progressively reduces force transmission above but not below the rigidity threshold.
Traction forces exerted by control cells on 5 kPa gels (blue) and 29 kPa gels (red) as a function of the efficiency of transfection with Talin1 Head L325R. Values are compared to mean forces of untransfected control cells (left) and talin shRNA cells (right). Note that in Fig. 1, talin 1 Head L325R data represent averages for well transfected cells only. Dotted lines represent sigmoidal fits to the data. (Control: 5 kPa, n = 35 cells; 29 kPa, n = 12 cells. Control + Talin 1 Head: 5 kPa, n = 37 cells; 29 kPa, n = 42 cells. Talin 2 shRNA: 5 kPa, n = 34 cells; 29 kPa, n = 29 cells). Data show 1 out of 3 independent experiments.
Supplementary Figure 6 Further quantifications of integrin and pFAK in adhesions.
(a) quantification of integrin density from staining images of ligand bound β3 for Control cells (red, n = 20, 31, 24, 33, 20, 29 fields respectively for increasing rigidity measured in 8–10 cells) and Talin 2 shRNA cells (blue, n = 20, 21, 25, 29, 24, 29 fields measured in 8–9 cells). Data show 1 out of 3 independent experiments. Integrin densities were significantly different between control and depleted cells only above 5 kPa (P < 0.001, two-way Anova). (b) Quantification of the percentage of cell spreading area covered by pFAK-positive adhesions from staining images of Control cells (red. n = 11, 10, 17, 17, 17, 15 cells respectively for increasing rigidity) and Talin 2 shRNA cells (blue, n = 11, 10, 10, 12, 10, 10 cells) as a function of substrate stiffness. Data show 1 out of 3 independent experiments. Significant differences were observed only above 5 kPa (P = 0.039, two-way Anova).
Supplementary Figure 7 Dependence of cell area and myosin phosphorylation on substrate stiffness.
(a) Quantification of cell area in response to substrate stiffness for control and talin 2 shRNA cells (Control: n = 17, 12, 35, 42, 42, 12 cells respectively for increasing stiffness; Talin 2 shRNA: n = 10, 11, 34, 23, 25, 29 cells). Data show 1 out of 14 independent experiments. Talin depletion did not have a significant effect (two-way Anova). (b) For cells plated on gels of the indicated stiffness, representative western blots of talin, GAPDH as loading control, phosphorylated myosin light chain and total myosin light chain for Control and Talin 2 shRNA cells. (c) Corresponding quantification of the phosphorylated/total myosin light chain ratio (pooled from n = 3 independent experiments). No significant differences were found (two-way Anova). (d) Representative western blots of phosphorylated myosin light chain and total myosin light chain for wild-type MEF cells. (e) Corresponding quantification of the phosphorylated/total myosin light chain ratio (pooled from n = 3 independent experiments). No significant differences were found, suggesting that myosin phosphorylation is not significantly affected in MEF cells regardless of talin (one-way Anova).
Supplementary Figure 8 Further analyses on the effects of vinculin fragments.
(a) Average forces in response to substrate stiffness for cells transfected with Talin 2 shRNA + VD1 (red, n = 10, 13, 11, 11, 13, 10 cells, respectively for increasing stiffness) and Talin 2 shRNA + VD1 A501 (blue, n = 11, 11, 12, 11, 11, 10 cells). No significant differences were found between transfections (two-way Anova). Data show 1 out of 3 independent experiments. (b) Quantification of Nuclear/Cytosolic YAP ratio for the same conditions as in (a) (Talin 2 shRNA + VD1:n = 26, 20, 30, 29, 32, 32 cells respectively for increasing stiffness; Talin 2 shRNA + VD1 A501: n = 22, 21, 21, 23, 23, 24 cells). No significant differences were found between transfections (two-way Anova). Data show 1 out of 3 independent experiments. (c) Quantification of Nuclear/Cytosolic YAP ratio for control cells transfected with VD1 (red) and or VD1 A501 (blue) as a function of transfection efficiency (measured as the relative intensity of EGFP fluorescence) on 29 kPa polyacrylamide gels (Control + VD1:n = 59 cells; Control + VD1 A501: n = 49 cells). Data show 1 out of 3 independent experiments. Dashed lines are a sigmoidal fit to the experimental results for each condition. Further confirming the blocking role of VD1, increasing transfection efficiencies progressively decreased nuclear localization of YAP. In contrast, increasing efficiencies of transfection with VD1 A501 had no effect.
Supplementary Figure 9 Fibronectin coating densities.
Resulting fibronectin coating densities on the surface of polyacrylamide gels of 5 and 29 kPa coated with solutions containing 1, 10, or 100 μg ml−1 of fibronectin. n = 6 gels in all cases except 100 μg ml−1–29 kPa (5 gels). Data pooled from two independent experiments.
Supplementary Figure 10 Unprocessed versions of the western blots shown in Supplementary Fig. 4.
(a,b) blots corresponding to panel b in Supplementary Fig. 4. (c) blots corresponding to panel d in Supplementary Fig. 4. All measured bands corresponded to the molecular weights of the different proteins as detailed by antibody providers: talin (225–235 kDa), GADPH (36 kDa), and MLC (18 kDa). Note that blots do not show the entire molecular weight spectrum because membranes were cut before antibody incubation to incubate each band only with the relevant antibody.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1057 kb)
41556_2016_BFncb3336_MOESM8_ESM.avi
Time-lapse of control cells transfected with lifeact-GFP and plated on fibronectin-coated substrates of increasing Young’s modulus (2-5-11-14-29 kPa from left to right). Scale bar is 20 μm. (AVI 1996 kb)
41556_2016_BFncb3336_MOESM9_ESM.avi
Time-lapse of talin 2 shRNA cells transfected with lifeact-GFP and plated on fibronectin-coated substrates of increasing Young’s modulus (2-5-11-14-29 kPa from left to right). Scale bar is 20 μm. (AVI 1733 kb)
Rights and permissions
About this article
Cite this article
Elosegui-Artola, A., Oria, R., Chen, Y. et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat Cell Biol 18, 540–548 (2016). https://doi.org/10.1038/ncb3336
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3336
This article is cited by
-
Adhesion energy controls lipid binding-mediated endocytosis
Nature Communications (2024)
-
Myosin-independent stiffness sensing by fibroblasts is regulated by the viscoelasticity of flowing actin
Communications Materials (2024)
-
The impact of tumor microenvironment: unraveling the role of physical cues in breast cancer progression
Cancer and Metastasis Reviews (2024)
-
Matrix viscoelasticity promotes liver cancer progression in the pre-cirrhotic liver
Nature (2024)
-
High expression of Talin-1 is associated with tumor progression and recurrence in melanoma skin cancer patients
BMC Cancer (2023)