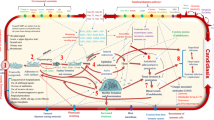
Abstract
Pathologies attributable to fungal infections represent a growing concern in both developed and developing countries. Initially discovered as opportunistic pathogens of immunocompromised hosts, fungi such as Candida albicans are now being placed at the centre of a more complex and dynamic picture in which the outcome of an infection is the result of an intricate network of molecular interactions between the fungus, the host and the commensal microflora co-inhabiting various host niches, and especially the gastrointestinal (GI) tract. The complexity of the host–fungal interaction begins with the numerous pathogen-associated molecular patterns (PAMPs) present on the fungal cell wall that are recognized by multiple pathogen-recognition receptors (PRRs), expressed by several types of host cells. PAMP–PRR interactions elicit a variety of intracellular signalling pathways leading to a wide array of immune responses, some of which promote fungal clearance while others contribute to pathogenesis. The picture is further complicated by the fact that numerous commensal bacteria normally co-inhabiting the host’s GI tract produce molecules that either directly modulate the survival and virulence of commensal fungi such as C. albicans or indirectly modulate the host’s antifungal immune responses. On top of this complexity, this host–microbiome–fungal interaction exhibits features of a dynamic system, in which the same fungi can easily switch between different morphological forms presenting different PAMPs at different moments of time. Furthermore, fungal pathogens can rapidly accumulate genomic alterations that further modify their recognition by the immune system, their virulence and their resistance to antifungal compounds. Thus, based on available molecular data alone, it is currently difficult to construct a coherent model able to explain the balance between commensalism and virulence and to predict the outcome of a fungal infection. Here, we review current advances in our understanding of this complex and dynamic system and propose new avenues of investigation to assemble a more complete picture of the host–fungal interaction, integrating microbiological and immunological data under the lens of systems biology and evolutionary genomics.

Similar content being viewed by others
References
Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133–63.
MacCallum DM. Candida infections and modelling disease. In: Ashbee R, Bignell EM, editors. Pathogenic yeasts. Berlin: Springer; 2010. p. 41–67.
Romani L, Fallarino F, De Luca A, Montagnoli C, D’Angelo C, Zelante T, Vacca C, Bistoni F, Fioretti MC, Grohmann U, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451(7175):211–5.
Singh N, Perfect JR. Immune reconstitution syndrome and exacerbation of infections after pregnancy. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2007;45(9):1192–9.
Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. J Microbiol. 2011;49(2):171–7.
Cottier F, Muhlschlegel FA. Sensing the environment: response of Candida albicans to the X factor. FEMS Microbiol Lett. 2009;295(1):1–9.
Casadevall A, Fang FC, Pirofski LA. Microbial virulence as an emergent property: consequences and opportunities. PLoS Pathog. 2011;7(7):e1002136.
Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6(1):67–78.
Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci USA. 2007;104(4):1366–70.
Rosas AL, Nosanchuk JD, Feldmesser M, Cox GM, McDade HC, Casadevall A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect Immun. 2000;68(5):2845–53.
Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11(4):275–88.
Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116(6):1642–50.
Miyazato A, Nakamura K, Yamamoto N, Mora-Montes HM, Tanaka M, Abe Y, Tanno D, Inden K, Gang X, Ishii K, et al. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect Immun. 2009;77(7):3056–64.
Jouault T, El Abed-El Behi M, Martinez-Esparza M, Breuilh L, Trinel PA, Chamaillard M, Trottein F, Poulain D. Specific recognition of Candida albicans by macrophages requires galectin-3 to discriminate Saccharomyces cerevisiae and needs association with TLR2 for signaling. J Immunol. 2006;177(7):4679–87.
Cambi A, Gijzen K, de Vries JM, Torensma R, Joosten B, Adema GJ, Netea MG, Kullberg BJ, Romani L, Figdor CG. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. Eur J Immunol. 2003;33(2):532–8.
Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol. 2008;10(10):2058–66.
Bittencourt VC, Figueiredo RT, da Silva RB, Mourao-Sa DS, Fernandez PL, Sassaki GL, Mulloy B, Bozza MT, Barreto-Bergter E. An alpha-glucan of Pseudallescheria boydii is involved in fungal phagocytosis and Toll-like receptor activation. J Biol Chem. 2006;281(32):22614–23.
Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–91.
Rizzetto L, Cavalieri D. Friend or foe: using systems biology to elucidate interactions between fungi and their hosts. Trends Microbiol. 2011;19(10):509–15.
Rizzetto L, Kuka M, De Filippo C, Cambi A, Netea MG, Beltrame L, Napolitani G, Torcia MG, D’Oro U, Cavalieri D. Differential IL-17 production and mannan recognition contribute to fungal pathogenicity and commensalism. J Immunol. 2010;184(8):4258–68.
Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van het Hoog M, Gordon P, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13(10):3452–65.
Zhao R, Daniels KJ, Lockhart SR, Yeater KM, Hoyer LL, Soll DR. Unique aspects of gene expression during Candida albicans mating and possible G(1) dependency. Eukaryot Cell. 2005;4(7):1175–90.
Staib P, Morschhauser J. Chlamydospore formation in Candida albicans and Candida dubliniensis—an enigmatic developmental programme. Mycoses. 2007;50(1):1–12.
Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8(1):31–8.
Drummond RA, Brown GD. The role of Dectin-1 in the host defence against fungal infections. Curr Opin Microbiol. 2011;14(4):392–9.
Dennehy KM, Brown GD. The role of the beta-glucan receptor Dectin-1 in control of fungal infection. J Leukoc Biol. 2007;82(2):253–8.
van de Veerdonk FL, Kullberg BJ, van der Meer JW, Gow NA, Netea MG. Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr Opin Microbiol. 2008;11(4):305–12.
Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459(7245):433–6.
Kumamoto CA. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14(4):386–91.
Eyerich K, Foerster S, Rombold S, Seidl HP, Behrendt H, Hofmann H, Ring J, Traidl-Hoffmann C. Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J Invest Dermatol. 2008;128(11):2640–5.
Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206(2):299–311.
Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332(6025):65–8.
Romani L, Puccetti P. Protective tolerance to fungi: the role of IL-10 and tryptophan catabolism. Trends Microbiol. 2006;14(4):183–9.
Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, Belladonna ML, Vacca C, Conte C, Mosci P, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. 2007;37(10):2695–706.
Bozza S, Zelante T, Moretti S, Bonifazi P, DeLuca A, D’Angelo C, Giovannini G, Garlanda C, Boon L, Bistoni F, et al. Lack of Toll IL-1R8 exacerbates Th17 cell responses in fungal infection. J Immunol. 2008;180(6):4022–31.
Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34(3):422–34.
Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48.
Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–8.
Neely AN, Law EJ, Holder IA. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun. 1986;52(1):200–4.
Adam B, Baillie GS, Douglas LJ. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol. 2002;51(4):344–9.
Gale D, Sandoval B. Response of mice to the inoculations of both Candida albicans and Escherichia coli. I. The enhancement phenomenon. J Bacteriol. 1957;73(5):616–24.
Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, Zhu Y, Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4(1):28–39.
Klaerner HG, Uknis ME, Acton RD, Dahlberg PS, Carlone-Jambor C, Dunn DL. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J Surg Res. 1997;70(2):161–5.
McAlester G, O’Gara F, Morrissey JP. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol. 2008;57(Pt 5):563–9.
Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2(1):27–36.
Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, et al. A bacterial cell–cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51(3):903–12.
Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC Jr, Mylonakis E. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105(38):14585–90.
Kim Y, Mylonakis E. Killing of Candida albicans filaments by Salmonella enterica serovar Typhimurium is mediated by sopB effectors, parts of a type III secretion system. Eukaryot Cell. 2011;10(6):782–90.
Fitzsimmons N, Berry DR. Inhibition of Candida albicans by Lactobacillus acidophilus: evidence for the involvement of a peroxidase system. Microbios. 1994;80(323):125–33.
Velraeds MM, van de Belt-Gritter B, van der Mei HC, Reid G, Busscher HJ. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J Med Microbiol. 1998;47(12):1081–5.
Huang CB, Alimova Y, Myers TM, Ebersole JL. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 2011;56(7):650–4.
Villena J, Salva S, Aguero G, Alvarez S. Immunomodulatory and protective effect of probiotic Lactobacillus casei against Candida albicans infection in malnourished mice. Microbiol Immunol. 2011;55(6):434–45.
Chen X, Xu J, Shuai J, Chen J, Zhang Z, Fang W. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int J Food Microbiol. 2007;115(3):307–12.
Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105(49):19474–9.
Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5.
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41.
Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67(7):3504–11.
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98.
Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–89.
Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10(3):260–72.
Reading NC, Kasper DL. The starting lineup: key microbial players in intestinal immunity and homeostasis. Front Microbiol. 2011;2:148.
Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9(4):176–80.
Yang F, Yan TH, Rustchenko E, Gao PH, Wang Y, Yan L, Cao YY, Wang QJ, Ji H, Cao YB, et al. High-frequency genetic contents variations in clinical Candida albicans isolates. Biol Pharm Bull. 2011;34(5):624–31.
Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468(7321):321–5.
Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135(5):879–93.
Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–70.
Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009;5(10):e1000705.
Hu G, Wang J, Choi J, Jung WH, Liu I, Litvintseva AP, Bicanic T, Aurora R, Mitchell TG, Perfect JR, et al. Variation in chromosome copy number influences the virulence of Cryptococcus neoformans and occurs in isolates from AIDS patients. BMC Genomics. 2011;12(1):526.
Selmecki A, Forche A, Berman J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell. 2010;9(7):991–1008.
Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio. 2011;2(4):e00129–11.
Forche A, Magee PT, Selmecki A, Berman J, May G. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics. 2009;182(3):799–811.
Dumitru R, Navarathna DH, Semighini CP, Elowsky CG, Dumitru RV, Dignard D, Whiteway M, Atkin AL, Nickerson KW. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot Cell. 2007;6(3):465–72.
Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460(7257):890–3.
Alby K, Bennett RJ. Interspecies pheromone signaling promotes biofilm formation and same-sex mating in Candida albicans. Proc Natl Acad Sci USA. 2011;108(6):2510–5.
Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22(10):2505–15.
Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6(5):e110.
Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010;22(6):809–15.
Berman J, Hadany L. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet. 2012. doi:10.1016/j.tig.2012.01.004.
Acknowledgments
Authors are grateful to Teresa Zelante, Judith Berman and Arturo Casadevall for critical reading of the manuscript. This work was supported by an A*STAR Investigatorship award to Norman Pavelka.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cottier, F., Pavelka, N. Complexity and dynamics of host–fungal interactions. Immunol Res 53, 127–135 (2012). https://doi.org/10.1007/s12026-012-8265-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-012-8265-y